mirror of
https://git.gfz-potsdam.de/naaice/tug.git
synced 2025-12-13 17:38:23 +01:00
3.8 KiB
3.8 KiB
2D Validation Examples
Simple setup using deSolve/ReacTran
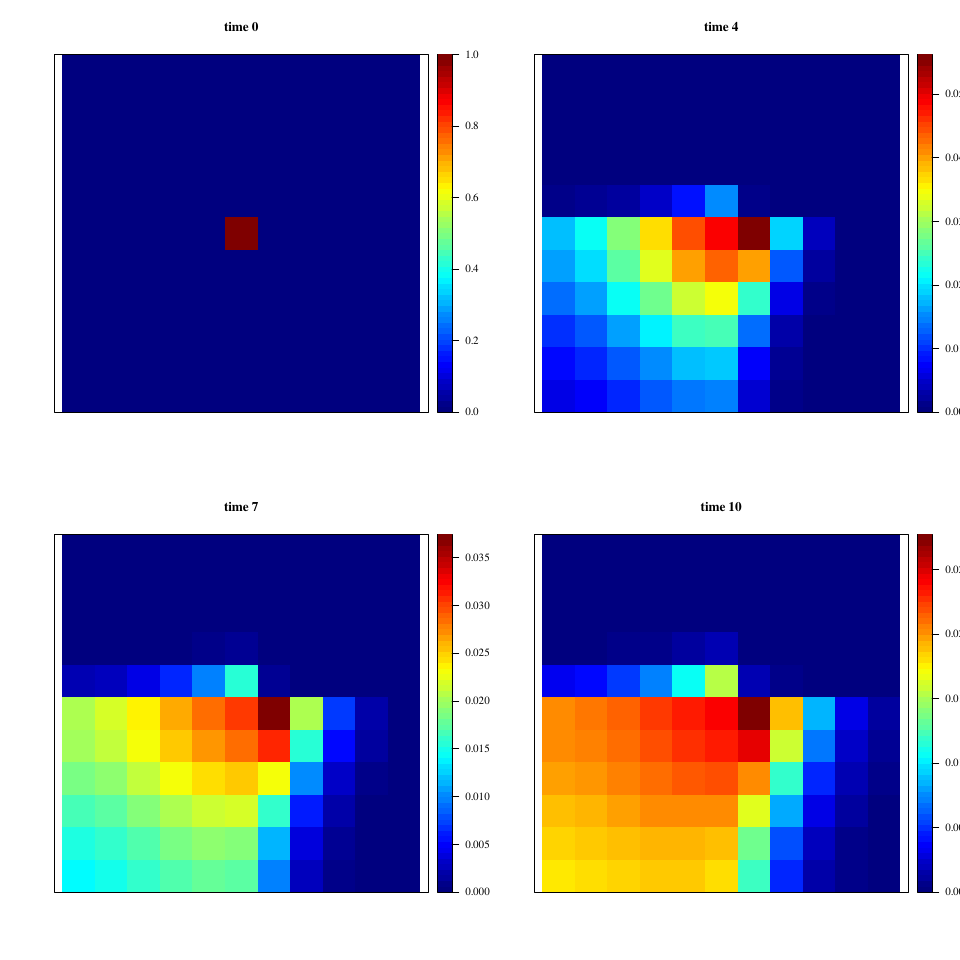
- Square of side 10
- Discretization: 11 × 11 cells
- All boundaries closed
- Initial state: 0 everywhere, 1 in the center (6,6)
- Time step: 1 s, 10 iterations
The matrix of spatially variable diffusion coefficients α is constant in 4 quadrants:
αx,y =
| 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
The relevant part of the R script used to produce these results is presented in listing 1; the whole script is at /max/tug/src/commit/788e59c4ef4746502b7d451e2c9294c0a5b18ef2/doc/scripts/HetDiff.R. A visualization of the output of the reference simulation is given in figure 1.
Note: all results from this script are stored in the outc matrix by
the deSolve function. I stored a different version into
/max/tug/src/commit/788e59c4ef4746502b7d451e2c9294c0a5b18ef2/scripts/gold/HetDiff1.csv: this file contains 11 columns (one
for each time step including initial conditions) and 121 rows, one for
each domain element, with no headers.
library(ReacTran)
library(deSolve)
## harmonic mean
harm <- function(x,y) {
if (length(x) != 1 || length(y) != 1)
stop("x & y have different lengths")
2/(1/x+1/y)
}
N <- 11 # number of grid cells
ini <- 1 # initial value at x=0
N2 <- ceiling(N/2)
L <- 10 # domain side
## Define diff.coeff per cell, in 4 quadrants
alphas <- matrix(0, N, N)
alphas[1:N2, 1:N2] <- 1
alphas[1:N2, seq(N2+1,N)] <- 0.1
alphas[seq(N2+1,N), 1:N2] <- 0.01
alphas[seq(N2+1,N), seq(N2+1,N)] <- 0.001
cmpharm <- function(x) {
y <- c(0, x, 0)
ret <- numeric(length(x)+1)
for (i in seq(2, length(y))) {
ret[i-1] <- harm(y[i], y[i-1])
}
ret
}
## Construction of the 2D grid
x.grid <- setup.grid.1D(x.up = 0, L = L, N = N)
y.grid <- setup.grid.1D(x.up = 0, L = L, N = N)
grid2D <- setup.grid.2D(x.grid, y.grid)
dx <- dy <- L/N
D.grid <- list()
## Diffusion coefs on x-interfaces
D.grid$x.int <- apply(alphas, 1, cmpharm)
## Diffusion coefs on y-interfaces
D.grid$y.int <- t(apply(alphas, 2, cmpharm))
# The model
Diff2Dc <- function(t, y, parms) {
CONC <- matrix(nrow = N, ncol = N, data = y)
dCONC <- tran.2D(CONC, dx = dx, dy = dy, D.grid = D.grid)$dC
return(list(dCONC))
}
## initial condition: 0 everywhere, except in central point
y <- matrix(nrow = N, ncol = N, data = 0)
y[N2, N2] <- ini # initial concentration in the central point...
## solve for 10 time units
times <- 0:10
outc <- ode.2D(y = y, func = Diff2Dc, t = times, parms = NULL,
dim = c(N, N), lrw = 1860000)