9.2 KiB
Numerical solution of diffusion equation in 2D with ADI Scheme
Finite differences with nodes as cells' centres
The 1D diffusion equation is:
\begin{align} \frac{\partial C }{\partial t} & = \frac{\partial}{\partial x} \left(\alpha \frac{\partial C }{\partial x} \right) \nonumber \\ & = \alpha \frac{\partial^2 C}{\partial x^2} \end{align}We aim at numerically solving eqn:1 on a spatial grid such as:
/max/tug/src/commit/e8b1c1daae4395dc29b36181cc8f1d8257a22ad7/doc/grid_pqc.pdf
The left boundary is defined on $x=0$ while the center of the first cell - which are the points constituting the finite difference nodes - is in $x=dx/2$, with $dx=L/n$.
The explicit FTCS scheme (as in PHREEQC)
We start by discretizing eqn:1 following an explicit Euler scheme and specifically a Forward Time, Centered Space finite difference.
For each cell index $i \in 1, \dots, n-1$ and assuming constant $\alpha$, we can write:
\begin{equation}\displaystyle \frac{C_ij+1 -C_ij}{Δ t} = α\frac{\frac{C^ji+1-C^ji}{Δ x}-\frac{C^ji-C^ji-1}{Δ x}}{Δ x}
\end{equation}
In practice, we evaluate the first derivatives of $C$ w.r.t. $x$ on the boundaries of each cell (i.e., $(C_{i+1}-C_i)/\Delta x$ on the right boundary of the i-th cell and $(C_{i}-C_{i-1})/\Delta x$ on its left cell boundary) and then repeat the differentiation to get the second derivative of $C$ on the the cell centre $i$.
This discretization works for all internal cells, but not for the domain boundaries ($i=0$ and $i=n$). To properly treat them, we need to account for the discrepancy in the discretization.
For the first (left) cell, whose center is at $x=dx/2$, we can evaluate the left gradient with the left boundary using such distance, calling $l$ the numerical value of a constant boundary condition:
\begin{equation}\displaystyle \frac{C_0j+1 -C_0j}{Δ t} = α\frac{\frac{C^j1-C^j0}{Δ x}- \frac{C^j0-l}{\frac{Δ x}{2}}}{Δ x}
\end{equation}
This expression, once developed, yields:
\begin{align}\displaystyle
C_0j+1 & = C_0j + \frac{α ⋅ Δ t}{Δ x^2} ⋅ ≤ft( C^j1-C^j0- 2 C^j0+2l \right) \nonumber
& = C_0j + \frac{α ⋅ Δ t}{Δ x^2} ⋅ ≤ft( C^j1- 3 C^j0 +2l \right)
\end{align}
In case of constant right boundary, the finite difference of point $C_n$ - calling $r$ the right boundary value - is:
\begin{equation}\displaystyle \frac{C_nj+1 -C_n^j}{Δ t} = α\frac{\frac{r - C^jn}{\frac{Δ x}{2}}- \frac{C^jn-C^jn-1}{Δ x}}{Δ x}
\end{equation}
Which, developed, gives
\begin{align}\displaystyle
C_nj+1 & = C_nj + \frac{α ⋅ Δ t}{Δ x^2} ⋅ ≤ft( 2 r - 2 C^jn -C^jn + C^jn-1 \right) \nonumber
& = C_nj + \frac{α ⋅ Δ t}{Δ x^2} ⋅ ≤ft( 2 r - 3 C^jn + C^jn-1 \right)
\end{align}
If on the right boundary we have closed or Neumann condition, the left derivative in eq. eqn:5 becomes zero and we are left with:
\begin{equation}\displaystyle C_nj+1 = C_nj + \frac{α ⋅ Δ t}{Δ x^2} ⋅ (C^jn-1 - C^j_n)
\end{equation}
A similar treatment can be applied to the BTCS implicit scheme.
Implicit BTCS scheme
First, we define the Backward Time difference:
\begin{equation} \frac{\partial C^{j+1} }{\partial t} = \frac{C^{j+1}_i - C^{j}_i}{\Delta t} \end{equation}Second the spatial derivative approximation, evaluated at time level $j+1$:
\begin{equation} \frac{\partial^2 C^{j+1} }{\partial x^2} = \frac{\frac{C^{j+1}_{i+1}-C^{j+1}_{i}}{\Delta x}-\frac{C^{j+1}_{i}-C^{j+1}_{i-1}}{\Delta x}}{\Delta x} \end{equation}Taking the 1D diffusion equation from eqn:1 and substituting each term by the equations given above leads to the following equation:
\begin{align}\displaystyle
\frac{C_ij+1 - C_ij}{Δ t} & = α\frac{\frac{Cj+1i+1-Cj+1i}{Δ x}-\frac{Cj+1i-Cj+1i-1}{Δ x}}{Δ x} \nonumber
& = α\frac{Cj+1i-1 - 2Cj+1i + Cj+1i+1}{Δ x^2}
\end{align}
This only applies to inlet cells with no ghost node as neighbor. For the left cell with its center at $\frac{dx}{2}$ and the constant concentration on the left ghost node called $l$ the equation goes as followed:
\begin{equation}\displaystyle \frac{C_0j+1 -C_0j}{Δ t} = α\frac{\frac{Cj+11-Cj+10}{Δ x}- \frac{Cj+10-l}{\frac{Δ x}{2}}}{Δ x}
\end{equation}
This expression, once developed, yields:
\begin{align}\displaystyle
C_0j+1 & = C_0j + \frac{α ⋅ Δ t}{Δ x^2} ⋅ ≤ft( Cj+11-Cj+10- 2 Cj+10+2l \right) \nonumber
& = C_0j + \frac{α ⋅ Δ t}{Δ x^2} ⋅ ≤ft( Cj+11- 3 Cj+10 +2l \right)
\end{align}
Now we define variable $s_x$ as followed:
\begin{equation} s_x = \frac{\alpha \cdot \Delta t}{\Delta x^2} \end{equation}Substituting with the new variable $s_x$ and reordering of terms leads to the equation applicable to our model:
\begin{equation}\displaystyle -C^j_0 = (2s_x) ⋅ l + (-1 - 3s_x) ⋅ Cj+1_0 + s_x ⋅ Cj+1_1
\end{equation}
The right boundary follows the same scheme. We now want to show the equation for the rightmost inlet cell $C_n$ with right boundary value $r$:
\begin{equation}\displaystyle \frac{C_nj+1 -C_nj}{Δ t} = α\frac{\frac{r-Cj+1n}{\frac{Δ x}{2}}- \frac{Cj+1n-Cj+1n-1}{Δ x}}{Δ x}
\end{equation}
This expression, once developed, yields:
\begin{align}\displaystyle
C_nj+1 & = C_nj + \frac{α ⋅ Δ t}{Δ x^2} ⋅ ≤ft( 2r - 2Cj+1n - Cj+1n + Cj+1n-1 \right) \nonumber
& = C_0j + \frac{α ⋅ Δ t}{Δ x^2} ⋅ ≤ft( 2r - 3Cj+1n + Cj+1n-1 \right)
\end{align}
Now rearrange terms and substituting with $s_x$ leads to:
\begin{equation}\displaystyle -C^j_n = s_x ⋅ Cj+1n-1 + (-1 - 3s_x) ⋅ Cj+1_n + (2s_x) ⋅ r
\end{equation}
TODO
- Tridiagonal matrix filling
Old stuff
Input
-
c$\rightarrow c$- containing current concentrations at each grid cell for species
- size: $N \times M$
- row-major
-
alpha$\rightarrow \alpha$- diffusion coefficient for both directions (x and y)
- size: $N \times M$
- row-major
-
boundary_condition$\rightarrow bc$- Defines closed or constant boundary condition for each grid cell
- size: $N \times M$
- row-major
Internals
-
A_matrix$\rightarrow A$- coefficient matrix for linear equation system implemented as sparse matrix
- size: $((N+2)\cdot M) \times ((N+2)\cdot M)$ (including ghost zones in x direction)
- column-major (not relevant)
-
b_vector$\rightarrow b$- right hand side of the linear equation system
- size: $(N+2) \cdot M$
- column-major (not relevant)
-
x_vector$\rightarrow x$- solutions of the linear equation system
- size: $(N+2) \cdot M$
- column-major (not relevant)
Calculation for $\frac{1}{2}$ timestep
Symbolic addressing of grid cells
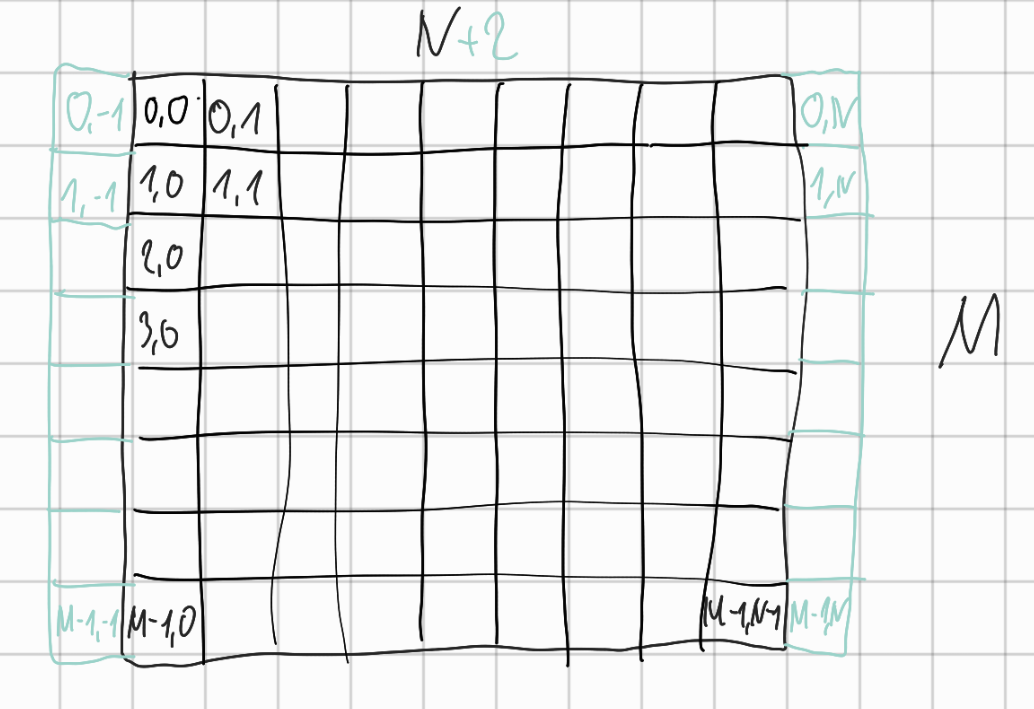
Filling of matrix $A$
- row-wise iterating with $i$ over
cand\alphamatrix respectively - addressing each element of a row with $j$
-
matrix $A$ also containing $+2$ ghost nodes for each row of input matrix $\alpha$
- $\rightarrow offset = N+2$
- addressing each object $(i,j)$ in matrix $A$ with $(offset \cdot i + j, offset \cdot i + j)$
Rules
$s_x(i,j) = \frac{\alpha(i,j)*\frac{t}{2}}{\Delta x^2}$ where $x$ defining the domain size in x direction.
For the sake of simplicity we assume that each row of the $A$ matrix is addressed correctly with the given offset.
Ghost nodes
$A(i,-1) = 1$
$A(i,N) = 1$
Inlet
$A(i,j) = \begin{cases} 1 & \text{if } bc(i,j) = \text{constant} \\ -1-2*s_x(i,j) & \text{else} \end{cases}$
$A(i,j\pm 1) = \begin{cases} 0 & \text{if } bc(i,j) = \text{constant} \\ s_x(i,j) & \text{else} \end{cases}$
Filling of vector $b$
- each elements assign a concrete value to the according value of the row of matrix $A$
-
Adressing would look like this: $(i,j) = b(i \cdot (N+2) + j)$
- $\rightarrow$ for simplicity we will write $b(i,j)$
Rules
Ghost nodes
$b(i,-1) = \begin{cases} 0 & \text{if } bc(i,0) = \text{constant} \\ c(i,0) & \text{else} \end{cases}$
$b(i,N) = \begin{cases} 0 & \text{if } bc(i,N-1) = \text{constant} \\ c(i,N-1) & \text{else} \end{cases}$
Inlet
$p(i,j) = \frac{\Delta t}{2}\alpha(i,j)\frac{c(i-1,j) - 2\cdot c(i,j) + c(i+1,j)}{\Delta x^2}$
\noindent $p$ is called t0_c inside code
$b(i,j) = \begin{cases} bc(i,j).\text{value} & \text{if } bc(i,N-1) = \text{constant} \\ -c(i,j)-p(i,j) & \text{else} \end{cases}$